QPCR troubleshooting involves identifying and correcting issues like low amplification efficiency or non-specific amplification. Key steps include optimizing primer design, reaction conditions, and template quality.
QPCR, or quantitative Polymerase Chain Reaction, is a powerful technique used to quantify DNA or RNA in a sample. Despite its effectiveness, several issues can arise during the process, such as low amplification efficiency, non-specific amplification, or inconsistent results. Addressing these problems involves careful optimization of various factors, including primer design, reaction conditions, and template quality.
Ensuring the use of high-quality reagents and maintaining a clean laboratory environment are also crucial. By systematically troubleshooting these aspects, you can achieve more reliable and accurate qPCR results.
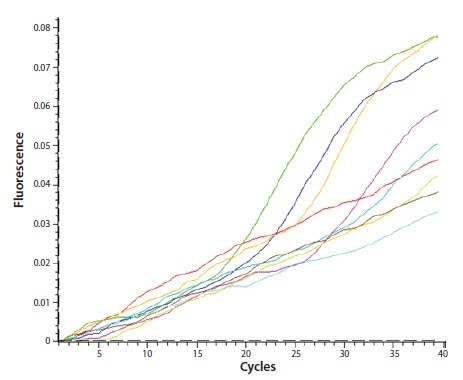
Credit: www.sigmaaldrich.com
Common Issues In Qpcr
Quantitative PCR (qPCR) is a powerful technique in molecular biology. It allows for the quantification of DNA and RNA. Despite its effectiveness, users often encounter common issues. These issues can affect the accuracy and reliability of qPCR results. Below, we discuss some of the most frequent problems and how to troubleshoot them.
Inconsistent Amplification
Inconsistent amplification is a frequent issue in qPCR experiments. It can lead to unreliable results. Several factors can cause this problem:
- Pipetting Errors: Ensure precise pipetting to avoid variability.
- Template Quality: Use high-quality DNA or RNA templates.
- Primer Design: Design primers specific to your target sequence.
- Master Mix Preparation: Prepare the master mix consistently.
To minimize pipetting errors, use calibrated pipettes. Always check the quality of your template with spectrophotometry. Ensure primers do not form dimers or secondary structures. Prepare the master mix in bulk to reduce variability.
High Background Signal
High background signal can obscure true amplification signals. This issue often results from:
- Contamination: Work in a clean environment to prevent contamination.
- Non-Specific Binding: Optimize annealing temperatures to reduce non-specific binding.
- Excessive Template: Use the optimal amount of template DNA or RNA.
To prevent contamination, use fresh reagents and wear gloves. Adjust the annealing temperature based on primer melting temperatures. Use just enough template to get clear, distinct peaks.
By addressing these common issues, you can improve the reliability of your qPCR results. Consistent practices and careful planning are key to successful qPCR experiments.
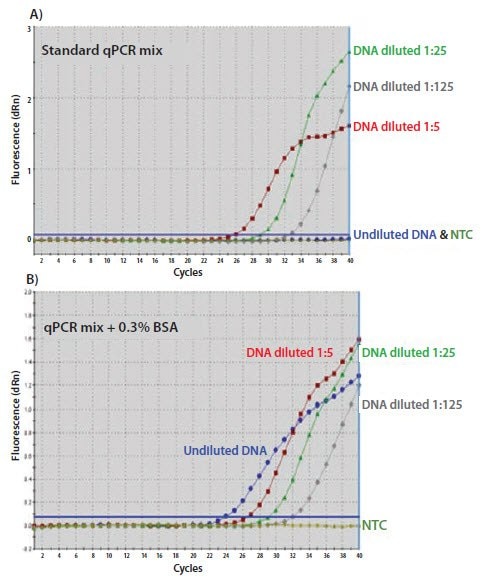
Credit: www.sigmaaldrich.com
Optimizing Primer Design
Optimizing primer design is a key step in qPCR troubleshooting. This ensures accurate and reliable results. Proper primer design can prevent many common qPCR issues. Below, we discuss essential aspects of primer design.
Choosing The Right Sequence
Choosing the right sequence for primers is crucial. Primers should be unique to your target region. This helps in specific binding. Use online tools to find the best sequences.
- Check for specificity using BLAST.
- Ensure the primer length is 18-22 bases.
- Aim for a GC content of 40-60%.
- Keep the melting temperature (Tm) around 60°C.
Avoiding Secondary Structures
Secondary structures in primers can cause inefficient binding. This leads to poor amplification. To avoid this, follow these guidelines:
- Avoid complementary sequences within primers.
- Use software tools to check for hairpins and dimers.
- Design primers without long runs of a single nucleotide.
| Factor | Recommendation |
|---|---|
| Primer Length | 18-22 bases |
| GC Content | 40-60% |
| Melting Temperature | Around 60°C |
By focusing on these aspects, you can enhance your qPCR results. Proper primer design is a cornerstone of successful experiments.
Sample Quality And Preparation
Ensuring the quality and proper preparation of your samples is crucial for successful qPCR. Poor sample quality can lead to unreliable results. Proper preparation helps in achieving accurate and reproducible data.
Dna/rna Purity
High DNA/RNA purity is essential for accurate qPCR results. Contaminants like proteins, phenol, and salts can inhibit the reaction. Use spectrophotometry to measure the purity of your samples.
A 260/280 ratio between 1.8 and 2.0 indicates pure DNA. For RNA, the ratio should be around 2.0. A lower ratio suggests protein contamination. A higher ratio indicates possible phenol contamination.
Consider using commercial kits for nucleic acid extraction. They are designed to provide high-purity samples.
Preventing Degradation
Sample degradation can compromise your qPCR results. RNA is particularly prone to degradation due to RNases.
Here are some tips to prevent degradation:
- Wear gloves to avoid introducing RNases.
- Use RNase-free reagents and consumables.
- Store samples at -80°C for long-term storage.
- Use stabilizing agents to protect RNA integrity.
For DNA, avoid repeated freeze-thaw cycles. Store DNA samples at -20°C or -80°C.
| Factor | DNA | RNA |
|---|---|---|
| Storage Temperature | -20°C to -80°C | -80°C |
| Contaminant Sensitivity | Low | High |
| Enzyme Sensitivity | Low | High (RNases) |
Reagent Quality And Storage
Proper reagent quality and storage are crucial for successful qPCR experiments. Poor storage can ruin reagents, causing unreliable results. Understanding proper storage and avoiding contamination can help maintain reagent integrity.
Proper Storage Conditions
Storing reagents properly ensures their longevity and performance. Always follow the manufacturer’s instructions.
- Enzymes should be stored at -20°C.
- DNA samples are best kept at -80°C.
- Primers and probes should be stored at -20°C.
Reagents should be kept in tightly sealed containers. This prevents evaporation and contamination. Avoid repeated freeze-thaw cycles to maintain reagent quality.
Avoiding Contamination
Contamination can ruin qPCR results. Here are some steps to avoid it:
- Use sterile pipette tips for each sample.
- Wear gloves to prevent skin oils from contaminating samples.
- Keep reagents in clean, dedicated areas.
Always use filtered tips to avoid aerosol contamination. Regularly clean work surfaces and equipment with 70% ethanol.
Thermocycler Settings
Proper thermocycler settings are crucial for successful qPCR experiments. Correct settings ensure accurate and reproducible results. Let’s explore key aspects of thermocycler settings.
Temperature Accuracy
The accuracy of the temperature settings is vital. Even a small deviation can lead to erroneous results.
- Calibration: Regular calibration of the thermocycler ensures precise temperature control.
- Validation: Use an external thermometer to validate the temperature settings.
- Thermal Uniformity: Ensure uniform temperature across all wells to avoid variations.
| Setting | Importance |
|---|---|
| Calibration | Ensures correct temperature |
| Validation | Confirms accurate settings |
| Thermal Uniformity | Prevents variability |
Cycle Optimization
Optimizing the cycling conditions can improve the efficiency of your qPCR.
- Annealing Temperature: Determine the optimal annealing temperature for your primers.
- Extension Time: Adjust the extension time based on the length of the amplicon.
- Number of Cycles: Use the appropriate number of cycles for your target.
Testing different conditions can help identify the best settings. This ensures high specificity and yield.
| Parameter | Recommendation |
|---|---|
| Annealing Temperature | Optimize for primers |
| Extension Time | Adjust for amplicon length |
| Number of Cycles | Use suitable cycles |
Data Analysis Techniques
Understanding data analysis techniques is crucial for accurate qPCR results. Proper analysis ensures reliable and reproducible data. In this section, we’ll cover two key techniques: threshold setting and standard curve calibration.
Threshold Setting
The threshold setting is a critical step in qPCR data analysis. It determines the point at which fluorescence rises above the background noise. Setting the threshold correctly helps identify the cycle threshold (Ct) value accurately.
Steps for Setting Threshold:
- Open your qPCR software.
- Locate the threshold setting option.
- Set the threshold just above the background signal.
- Verify the threshold placement across all samples.
Tips for Accurate Threshold Setting:
- Avoid placing the threshold too high.
- Ensure consistency across different runs.
- Adjust based on specific assay requirements.
Standard Curve Calibration
Standard curve calibration is essential for quantifying unknown samples. It helps in determining the efficiency of the qPCR reaction.
Creating a Standard Curve:
- Prepare a series of known concentrations.
- Run these standards in your qPCR experiment.
- Plot the Ct values against the log of the concentrations.
- Generate the standard curve using the software.
Evaluating the Standard Curve:
- Check the R2 value; it should be close to 1.
- Ensure the slope is between -3.3 and -3.6.
- Verify the efficiency is between 90% and 110%.
Controls And Replicates
Effective qPCR troubleshooting relies on understanding controls and replicates. These elements ensure the reliability and accuracy of your results. Using appropriate controls and replicates helps identify issues and validate findings.
Positive And Negative Controls
Positive controls confirm that the qPCR reaction works. They contain a known target sequence that should amplify. A positive control ensures the reagents and equipment are functioning. If the positive control fails, check the reagents and equipment.
Negative controls ensure no contamination in the reagents. They contain all components except the target sequence. If a negative control shows amplification, contamination might be present. Use separate areas for setup and processing to avoid contamination.
Technical And Biological Replicates
Technical replicates involve repeating the same sample multiple times. This helps identify pipetting errors and variability in the qPCR process. Use at least three technical replicates for reliable data. Consistent results across replicates indicate precision in your technique.
Biological replicates involve using different samples from the same group. This accounts for natural variability between biological samples. Use at least three biological replicates to ensure results reflect real biological differences. Biological replicates help generalize findings across a population.
| Type of Control | Purpose |
|---|---|
| Positive Control | Confirms qPCR reaction and reagent functionality |
| Negative Control | Checks for contamination |
| Type of Replicate | Purpose |
|---|---|
| Technical Replicates | Identify pipetting errors and qPCR variability |
| Biological Replicates | Account for natural sample variability |
Troubleshooting Guides
Real-time PCR, or qPCR, is a powerful technique used in molecular biology. Sometimes things go wrong. This troubleshooting guide helps you fix common qPCR issues.
Error Identification
Identifying errors early saves time and resources. Here are common qPCR errors:
- No amplification
- Low efficiency
- High background noise
- Inconsistent results
Step-by-step Solutions
Follow these steps to solve your qPCR issues:
No Amplification
- Check reagent quality and expiration dates.
- Verify template DNA quality and quantity.
- Ensure primer specificity and concentration.
Low Efficiency
- Optimize primer design.
- Check for inhibitors in the template.
- Adjust annealing temperature.
High Background Noise
- Use high-quality reagents.
- Reduce primer-dimer formation.
- Optimize cycling conditions.
Inconsistent Results
- Ensure consistent pipetting techniques.
- Use fresh reagents for each run.
- Standardize template and primer concentrations.

Credit: www.idtdna.com
Frequently Asked Questions
What Can Go Wrong In Qpcr?
Contamination can cause inaccurate results. Poor primer design affects specificity. Inconsistent sample preparation leads to variability. Inefficient reagents result in weak signals. Incorrect thermal cycling settings impact amplification.
Why Didn’t My Qpcr Work?
Your qPCR might have failed due to poor sample quality, incorrect primer design, or reagent issues. Ensure proper thermal cycling conditions and accurate pipetting. Check for contamination and verify instrument calibration.
What Are The Sources Of Error In Qpcr?
Sources of error in qPCR include pipetting inaccuracies, primer-dimer formations, suboptimal reaction conditions, and sample contamination. Inconsistent RNA quality and reverse transcription efficiency also affect results.
How To Troubleshoot Real-time Pcr?
To troubleshoot real-time PCR, check reagent quality, verify primer specificity, and optimize annealing temperatures. Ensure proper sample preparation and calibrate the thermal cycler. Regularly inspect equipment for maintenance. Use positive and negative controls to identify issues.
Conclusion
Effective qPCR troubleshooting is essential for reliable results. Follow best practices and maintain consistent protocols. Regularly calibrate your equipment and optimize reagents. Stay vigilant for common issues and address them promptly. By doing so, you ensure accurate and reproducible data, enhancing your research outcomes.